Cerebral 2.8.2 / CerebralWeb 1.0
:: DESCRIPTION
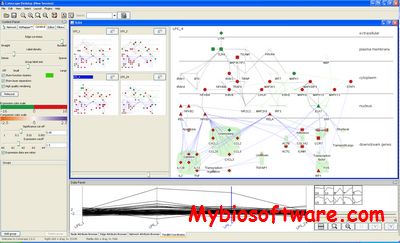
Cerebral (Cell Region-Based Rendering And Layout) is a Java plugin for the widely used open-source Cytoscape molecular interaction viewer.
CerebralWeb is a light-weight JavaScript plug-in that extends Cytoscape.js to enable fast and interactive visualisation of molecular interaction networks stratified based on subcellular localisation or other custom annotation.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/Linux/ MacOsX
- Java
- Cytoscape
:: DOWNLOAD
:: MORE INFORMATION
Citation
Barsky A, Gardy JL, Hancock REW, and Munzner T. (2007)
Cerebral: a Cytoscape plugin for layout of and interaction with biological networks using subcellular localization annotation.
Bioinformatics 23(8):1040-2.
Database (Oxford). 2015 May 7;2015. pii: bav041. doi: 10.1093/database/bav041. Print 2015.
CerebralWeb: a Cytoscape.js plug-in to visualize networks stratified by subcellular localization.
Frias S1, Bryan K1, Brinkman FS1, Lynn DJ