XRNA 2.0
:: DESCRIPTION
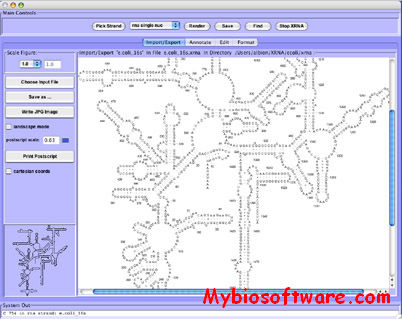
XRNA is a Java based suite of tools for the creation, annotation and display of RNA secondary structure diagrams.XRNA provides editing tools for easy modification of publication quality secondary diagrams that can be either drawn manually, or through automatic generation. Other features include grouping, numbering and structure annotation. XRNA secondary structures may be saved in a native format, or exported as postscript for printing and further manipulation in programs such as Adobe Illustrator.
::DEVELOPER
RNA Center, UCSC
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / MacOsX / Linux
- Perl
:: DOWNLOAD
:: MORE INFORMATION