CREATE 1.37
:: DESCRIPTION
CREATE is software for the creation of new and conversion of existing data input files for 64 genetic data analysis software programs. Programs are grouped into areas of sibship reconstruction, parentage assignment, effective population size, quantitative genetics, general genetic data analysis, and specialized genetic applications.
::DEVELOPER
Jason Coombs ,jcoombs@cns.umass.edu ,Ben Letcher, and Keith Nislow
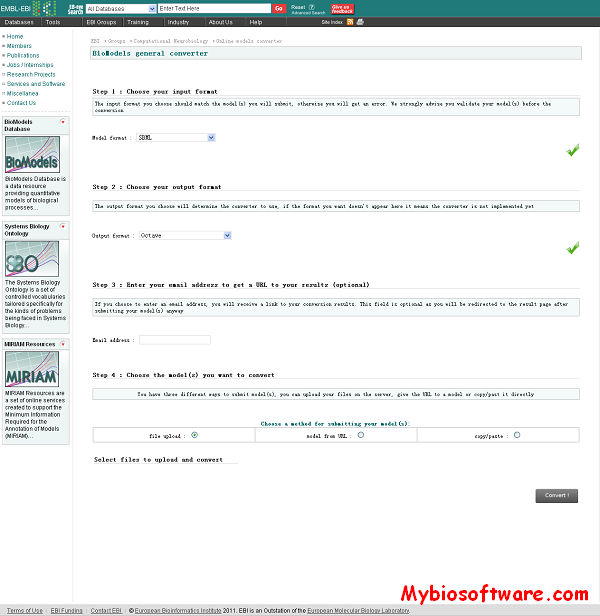
:: SCREENSHOTS
:: REQUIREMENTS
- Windows
:: DOWNLOAD
:: MORE INFORMATION
Citation
create: a software to create input files from diploid genotypic data for 52 genetic software programs.
Coombs JA, Letcher BH, Nislow KH.
Mol Ecol Resour. 2008 May;8(3):578-80. doi: 10.1111/j.1471-8286.2007.02036.x.