CRC 1.1
:: DESCRIPTION
CRC (Chinese Restaurant Cluster) implements a model-based Bayesian clustering algorithm. The cluster assignment procedure can be regarded as following a iterative Chinese restaurant process. This program is designed to cluster microarray gene expression data collected from multiple experiments. missing data is allowed. The program is written in C++, and can be run under Linux, Unix, Windows, MAC OSX operating system as a command line exexutable. CRC has the following features comparing to other clustering tools: 1) able to infer number of clusters, 2) able to cluster genes displaying time-shifted and/or inverted correlations, 3) able to tolerate missing genotype data and 4) provide confidence measure for clusters generated. Here is some more details on why you should try CRC for your microarray data analysis.
::DEVELOPER
Steve Qin @ the Center for Statistical Genetics
:: SCREENSHOTS
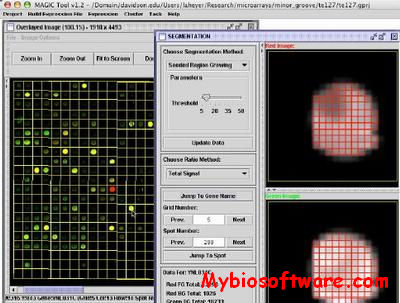
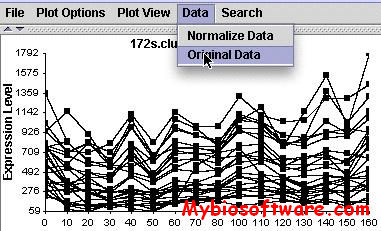
N/A
:: REQUIREMENTS
- Linux / Windows / MacOsX
:: DOWNLOAD
:: MORE INFORMATION
Citation
Qin ZS.
Clustering microarray gene expression data using weighted Chinese restaurant process.
Bioinformatics. 2006 Aug 15;22(16):1988-97. Epub 2006 Jun 9.