diffReps 1.55.6
:: DESCRIPTION
diffReps is developed to find different peaks in ChIP-seq. It scans the whole genome using a sliding window, performing millions of statistical tests and report the significant hits. diffReps takes into account the biological variations within a group of samples and uses that information to enhance the statistical power. Considering biological variation is of high importance, especiallly for in vivo brain tissues.
::DEVELOPER
:: SCREENSHOTS
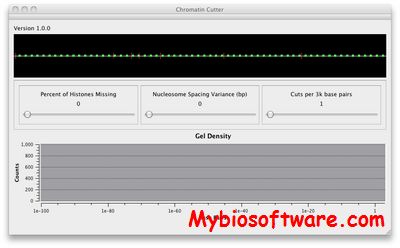
N/A
:: REQUIREMENTS
- Linux
- Perl
:: DOWNLOAD
:: MORE INFORMATION
Citation
PLoS One. 2013 Jun 10;8(6):e65598. doi: 10.1371/journal.pone.0065598. Print 2013.
diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates.
Shen L, Shao NY, Liu X, Maze I, Feng J, Nestler EJ.