CASSys
:: DESCRIPTION
CASSys (ChIP-seq data Analysis Software System) is an integrated system that supports all steps of computational ChIP-seq data analysis and supersedes the laborious application of several single command line tools. CASSys provides functionality ranging from quality assessment and -control of short reads, over the mapping of reads against a reference genome (readmapping) and the detection of enriched regions (peakdetection) to various follow-up analyses. The latter are accessible via a state-of-the-art web interface and can be performed interactively by the user. The follow-up analyses allow for flexible user defined association of putative interaction sites with genes, visualization of their genomic context with an integrated genome browser, the detection of putative binding motifs, the identification of over-represented Gene Ontology-terms, pathway analysis and the visualization of interaction networks. The system is client-server based, accessible via a web browser and does not require any software installation on the client side.
CASSys Online Server
::DEVELOPER
Junior Research Group for Application-oriented Bioinformatics, ,Center for Bioinformatics, University of Hamburg
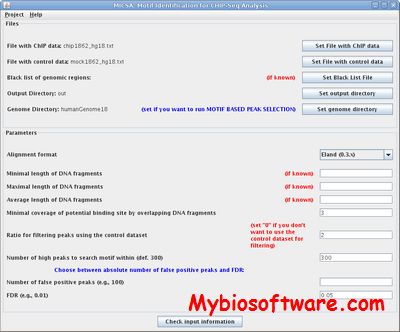
:: SCREENSHOTS

:: REQUIREMENTS
:: DOWNLOAD
 NO
NO
:: MORE INFORMATION
Citation:
Alawi, M., Kurtz, S. Beckstette, M. (2011).
CASSys: an integrated software system for the interactive analysis of ChIP-seq data.
Journal of Integrative Bioinformatics, 8(2):155,2011.