smoothseg 0.0.4
:: DESCRIPTION
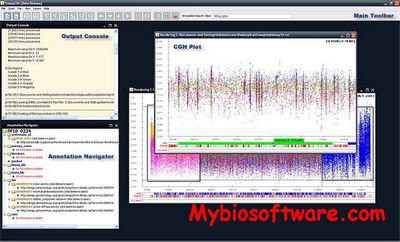
smoothseg is an R package to compute smooth-segmentation of array CGH data, including the estimation of FDR for comparative studies.
::DEVELOPER
Huang Jian <j.huang@ucc.ie>, Prof. Yudi Pawitan ,Arief Gusnanto <Arief.Gusnanto@mrc-bsu.cam.ac.uk>
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Windows/Linux /MacOsX
- R package
:: DOWNLOAD
:: MORE INFORMATION
Citation
Bioinformatics. 2007 Sep 15;23(18):2463-9. Epub 2007 Jul 27.
Robust smooth segmentation approach for array CGH data analysis.
Huang J1, Gusnanto A, O’Sullivan K, Staaf J, Borg A, Pawitan Y.