CGAT
:: DESCRIPTION
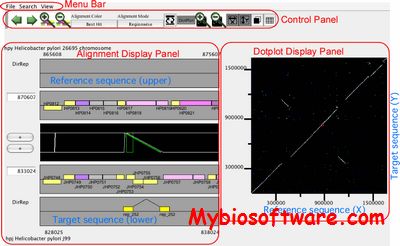
CGAT is a Comparative Genome Analysis Tool that was depeloped for detailed comparison of closely related bacterial-sized genomes. CGAT visualizes precomputed pairwise genome alignments on both dotplot and alignment viewers. Users can put several information on this alignment, such as existence of tandem repeats or interspersed repetitive sequences and changes in codon usage bias, to facilitate interpretation of the observed genomic changes.
::DEVELOPER
Ikuo Uchiyama (uchiyama@nibb.ac.jp)
:: SCREENSHOTS
:: REQUIREMENTS
- Linux / Windows/ MacOsX
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
CGAT: a comparative genome analysis tool for visualizing alignments in the analysis of complex evolutionary changes between closely related genomes.
Uchiyama I, Higuchi T, Kobayashi I.
BMC Bioinformatics. 2006 Oct 24;7:472.