CAT/BAT v5.2.3
:: DESCRIPTION
Contig Annotation Tool (CAT) and Bin Annotation Tool (BAT) are pipelines for the taxonomic classification of long DNA sequences and metagenome assembled genomes (MAGs/bins) of both known and (highly) unknown microorganisms, as generated by contemporary metagenomics studies.
::DEVELOPER
:: SCREENSHOTS
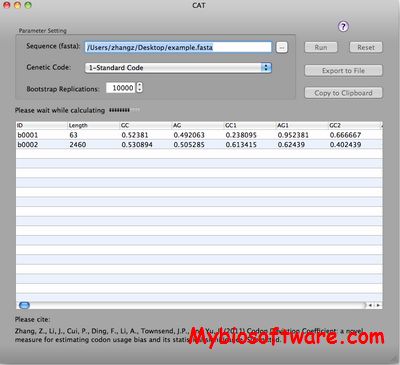
N/A
:: REQUIREMENTS
- Linux
- Python
:: DOWNLOAD
:: MORE INFORMATION
Citation
von Meijenfeldt FAB, Arkhipova K, Cambuy DD, Coutinho FH, Dutilh BE.
Robust taxonomic classification of uncharted microbial sequences and bins with CAT and BAT.
Genome Biol. 2019 Oct 22;20(1):217. doi: 10.1186/s13059-019-1817-x. PMID: 31640809; PMCID: PMC6805573.