PRESTO 1.01
:: DESCRIPTION
PRESTO is optimized software for permutation testing and for computing empirical distributions of order statistics via permutation. PRESTO is extremely fast and can efficiently analyze data from one and two-stage whole genome association studies with millions of markers genotyped on thousands of samples.
::DEVELOPER
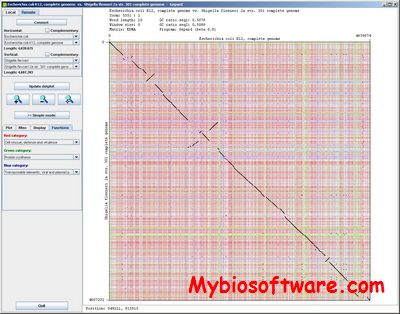
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux / Windows / MacOsX
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation:
B L Browning (2008)
PRESTO: Rapid calculation of order statistic distributions and multiple-testing adjusted P-values via permutation for one and two-stage genetic association studies.
BMC Bioinformatics 9:309.