Bioclojure
:: DESCRIPTION
BioClojure is an open-source library for the manipulation of biological sequence data written in the language Clojure. BioClojure aims to provide a functional framework for the processing of biological sequence data that provides simple mechanisms for concurrency and lazy evaluation of large datasets.
::DEVELOPER
:: SCREENSHOTS
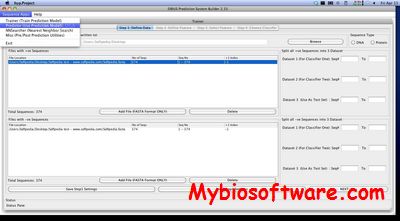
N/A
:: REQUIREMENTS
- Windows/Linux/MacOsX
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Bioclojure: a functional library for the manipulation of biological sequences.
Plieskatt J, Rinaldi G, Brindley PJ, Jia X, Potriquet J, Bethony J, Mulvenna J.
Bioinformatics. 2014 May 2. pii: btu311