GEDEVO / GEDEVO-M / CytoGEDEVO 1.0.3.1
:: DESCRIPTION
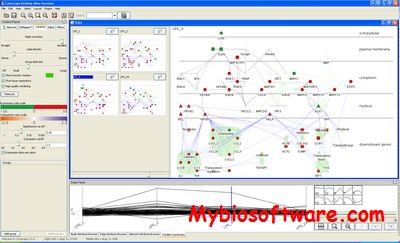
GEDEVO (Graph Edit Distance + EVOlution) is a software tool for solving the network alignment problem.
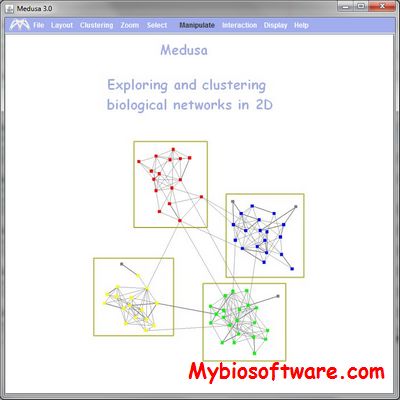
GEDEVO-M is an extension of the idea implemented in GEDEVO to multiple input networks.
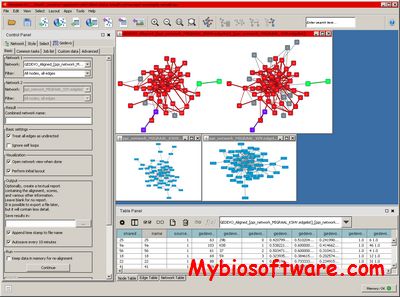
CytoGEDEVO is a Cytoscape plugin that implements the GEDEVO method for pairwise global network alignment.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/Linux / MacOsX
- Java
- Cytoscape
:: DOWNLOAD
 GEDEVO / GEDEVO-M / CytoGEDEVO
GEDEVO / GEDEVO-M / CytoGEDEVO
:: MORE INFORMATION
Citation
Rashid Ibragimov, Maximilian Malek, Jiong Guo, Jan Baumbach:
GEDEVO: An Evolutionary Graph Edit Distance Algorithm for Biological Network Alignment.
GCB 2013:68-79
CytoGEDEVO – Global alignment of biological networks with Cytoscape.
Malek M, Ibragimov R, Albrecht M, Baumbach J.
Bioinformatics. 2015 Dec 14. pii: btv732