MBSTAR
:: DESCRIPTION
MBSTAR (Multiple instance learning of Binding Sites of miRNA TARgets) is a web server for accurate prediction of true or functional miRNA binding sites.
::DEVELOPER
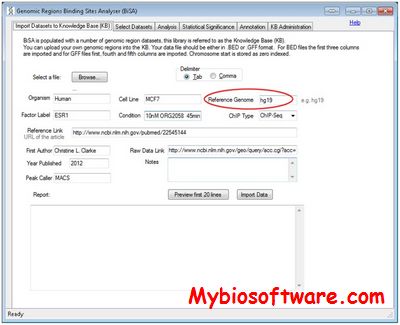
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux
:: DOWNLOAD
:: MORE INFORMATION
Citation
Sci Rep. 2015 Jan 23;5:8004. doi: 10.1038/srep08004.
MBSTAR: multiple instance learning for predicting specific functional binding sites in microRNA targets.
Bandyopadhyay S, Ghosh D, Mitra R, Zhao Z