BamView 1.2.11
:: DESCRIPTION
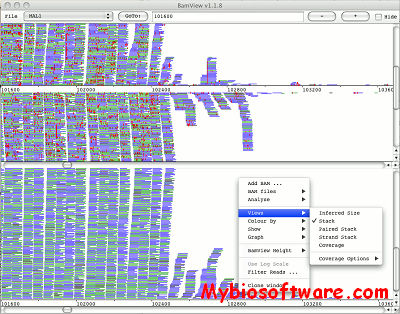
BamView is a free interactive display of read alignments in BAM data files.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / MacOsX / Linux
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Brief Bioinform. 2013 Mar;14(2):203-12. doi: 10.1093/bib/bbr073. Epub 2012 Jan 16.
BamView: visualizing and interpretation of next-generation sequencing read alignments.
Carver T1, Harris SR, Otto TD, Berriman M, Parkhill J, McQuillan JA.
Tim Carver; Ulrike Böhme, Thomas D. Otto; Julian Parkhill; Matthew Berriman
BamView: Viewing mapped read alignment data in the context of the reference sequence.
Bioinformatics 2010;