Tilescope 2.0
:: DESCRIPTION
Tilescope is a fully integrated data processing pipeline for analyzing tiling array data. In a completely automated fashion, it will normalize signals between channels and across arrays, combine replicate experiments, score each array element, and identify genomic features.
::DEVELOPER
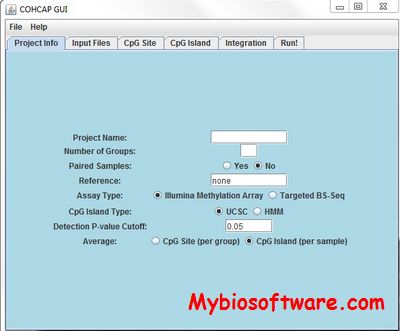
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux /Windows / MacOsX
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Genome Biol. 2007;8(5):R81.
Tilescope: online analysis pipeline for high-density tiling microarray data.
Zhang ZD1, Rozowsky J, Lam HY, Du J, Snyder M, Gerstein M.