PROBCONS 1.12
:: DESCRIPTION
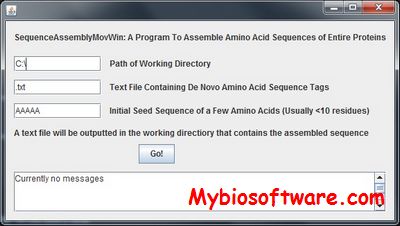
PROBCONS is an efficient protein multiple sequence alignment program, which has demonstrated a statistically significant improvement in accuracy compared to several leading alignment tools.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux
:: DOWNLOAD
:: MORE INFORMATION
Citation
Do, C.B., Mahabhashyam, M.S.P., Brudno, M., and Batzoglou, S. 2005.
PROBCONS: Probabilistic Consistency-based Multiple Sequence Alignment.
Genome Research 15: 330-340.