ASpipe 2.1.8
:: DESCRIPTION
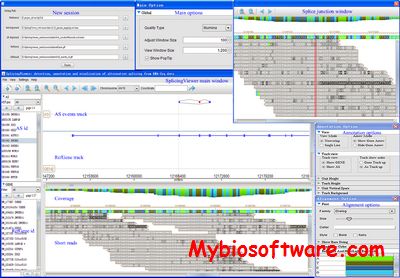
ASpipe (alternative splicing) is a pipeline to process GeneSeqer/GMAP alignments and identify alternative splicing (AS) events from the alignments.ASpipe extracts coordinates and scores for high-quality intron/exon/alignments from the original program outputs and stores them in MySQL5.0 databases.
::DEVELOPER
The Brendel Group @ Indiana University
:: REQUIREMENTS
:: DOWNLOAD
:: MORE INFORMATION
Citation
Wang, B.-B., O’Toole, M., Brendel, V. & Young, N.D. (2008)
Cross-species EST alignments reveal novel and conserved alternative splicing events in legumes.
BMC Plant Biol. 8:17.