SWAP 0.9
:: DESCRIPTION
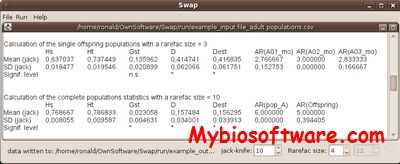
SWAP was developed to calculate the genetic diversity (AR, Hs, Ht) of and the genetic differentiation (Gst, D, Dest) among different half-sib families (embryos, respective pollen clouds) and within and among two different generations. Calculations are based on the father alleles. In the case that the embryo has the same genotype as the mother, it is not possible to decide which allele comes from the mother and which one comes from the father. In such ambiguous cases, the genetic parameters are measured via a Jackknifing-procedure. In addition, SWAP calculates all genetic parameters within and among several adult populations.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / Linux / MacOSX
- Java
:: DOWNLOAD
:: MORE INFORMATION