SNAVI 20081120
:: DESCRIPTION
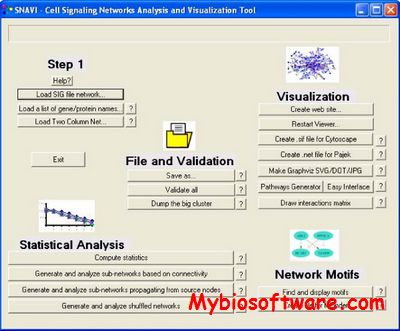
SNAVI (Signaling Networks Analysis and Visualization) is a Windows-based desktop application that implements standard network analysis methods to compute the clustering, connectivity distribution, and detection of network motifs, as well as provides means to visualize networks and network motifs. SNAVI is capable of generating linked web pages from network datasets loaded in text format. SNAVI can also create networks from lists of gene or protein names. SNAVI is a useful tool for analyzing, visualizing and sharing cell signaling data. SNAVI is open source free software.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows
:: DOWNLOAD
:: MORE INFORMATION
Citation
BMC Syst Biol. 2009 Jan 20;3:10. doi: 10.1186/1752-0509-3-10.
SNAVI: Desktop application for analysis and visualization of large-scale signaling networks.
Ma’ayan A, Jenkins SL, Webb RL, Berger SI, Purushothaman SP, Abul-Husn NS, Posner JM, Flores T, Iyengar R.