ENIGMA 1.1
:: DESCRIPTION
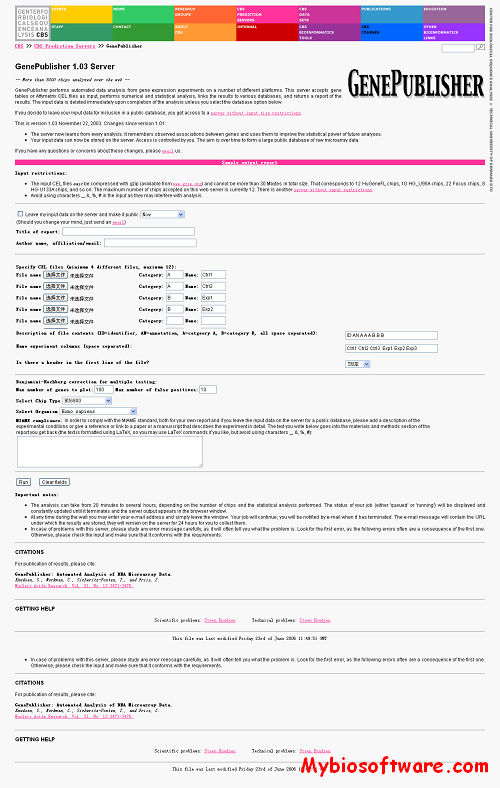
ENIGMA – Expression Network Inference and Global Module Analysis. It is a software tool to analyze the modular structure of perturbational gene expression data, based on the use of combinatorial statistics and graph-based clustering. The modules are further characterized by incorporating other data types, e.g. GO annotation, protein interactions and transcription factor binding information, and by suggesting regulators that might have an effect on the expression of (some of) the genes in the module.
::DEVELOPER
VIB (Flanders Interuniversitary Institute for Biotechnology)
Bioinformatics and Evolutionary Genomics Research Group at Ghent University
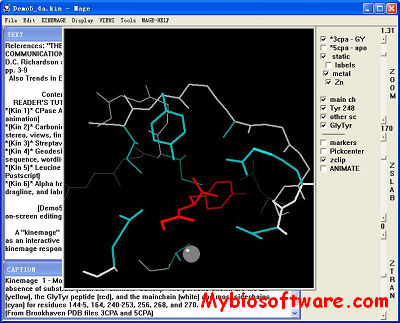
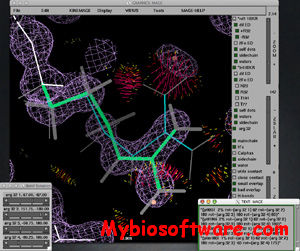
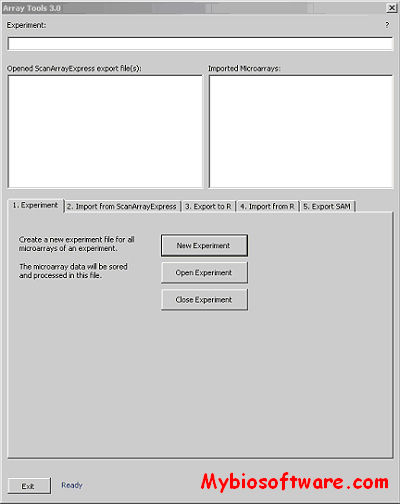
:: SCREENSHOTS
Command Line Tool
:: REQUIREMENTS
:: DOWNLOAD
:: MORE INFORMATION
If you use ENIGMA in your research, please cite:
Steven Maere, Patrick Van Dijck, Martin Kuiper (2008) Extracting expression modules from perturbational gene expression compendia. BMC Systems Biology 2:33 (PubMed, BMC)