mVISTA
:: DESCRIPTION
mVISTA is a set of programs for comparing DNA sequences from two or more species up to megabases long and visualize these alignments with annotation information. mVISTA has a clean output, allowing for easy identification of sequence similarities and differences, and is easily configurable, enabling the visualization of alignments of various lengths at different levels of resolution. It is implemented as an on-line server that provides access to global pairwise, multiple and glocal (global with rearrangements) alignment tools.
::DEVELOPER
Genomics Division of Lawrence Berkeley National Laboratory
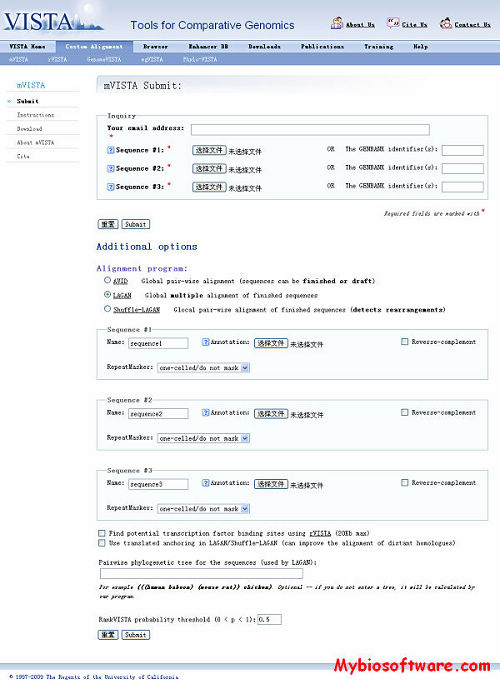
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / Mac OsX / Linux
- JAVA
:: DOWNLOAD
:: MORE INFORMATION
mVISTA and Avid are free for academic or non-profit research institutions to use for internal research purposes. Commercial companies may submit sequences over the Web using the VISTA web site at no charge. To obtain a copy of mVISTA or Avid for internal use, commercial entities are required to purchase a site license for commercial use.
Citation:
Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W273-9
Mayor C., Brudno M., Schwartz J. R., Poliakov A., Rubin E. M., Frazer K. A., Pachter L. S. and Dubchak I. (2000) VISTA: Visualizing Global DNA Sequence Alignments of Arbitrary Length. Bioinformatics, 16:1046.