miRDeep2 0.1.3 / miRDeep* 3.8
:: DESCRIPTION
miRDeep is a software package for identification of novel and known miRNAs in deep sequencing data. Furthermore, it can be used for miRNA expression profiling across samples. Last, a new module for preprocessing of raw Illumina sequencing data produces files for downstream analysis with the miRDeep2 or quantifier module.
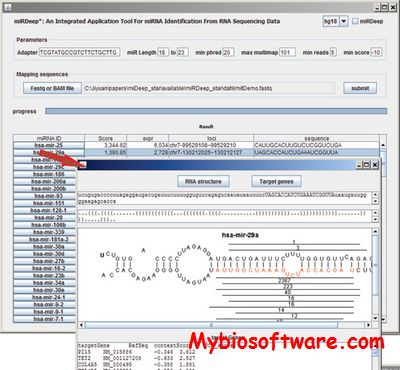
miRDeep* is an integrated miRNA identification tool, which is modeled off miRDeep, but the precision of detecting novel miRNAs is improved by introducing new strategies to identify precursor miRNAs. MiRDeep* has a user-friendly graphic interface and accepts raw data in FastQ and SAM/BAM format.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / Linux / MacOsX
- Perl / java
:: DOWNLOAD
:: MORE INFORMATION
Citation
An, J., Lai, J., Lehman, M.L. and Nelson, C.C. (2013)
miRDeep*: an integrated application tool for miRNA identification from RNA sequencing data.
Nucleic Acids Res, 41, 727-737.
Friedländer MR., Chen, W., Adamidi, C., Maaskola, J., Einspanier, R., Knespel, S., Rajewsky, N.
‘Discovering microRNAs from deep sequencing data using miRDeep‘,
Nature Biotechnology, 26, 407-415 (2008)