LOHAS 2.3
:: DESCRIPTION
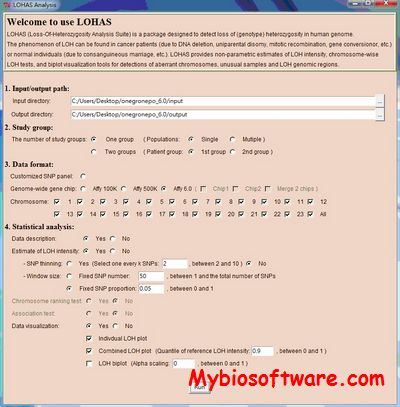
LOHAS (Loss-Of-Heterozygosity Analysis Suite) written in R and R GUI is developed to estimating homozygous intensity, identifying samples with unusual genomes, clustering samples with close LOH structures, and mapping the genomic segments bearing LOH by analyzing data of genome-wide SNP arrays or customized SNP arrays. In addition to cancer genetics and cancer genomics, LOHAS can also be applied to study long contiguous stretches of homozygosity (LCSH) in general populations. The LCSH analysis aids to identify samples with complex LCSH patterns, separate samples with different genetic background and gender in genetics, and locate LCSH regions.
::DEVELOPER
:: SCREENSHOTS
::REQUIREMENTS
:: DOWNLOAD
:: MORE INFORMATION
Citation
Yang, H.-C., Chang, L.-C., Huggins, R. M., Chen, C.-H. and Mullighan, C. G. (2011/05).
LOHAS: Loss-of-heterozygosity analysis suite.
Genetic Epidemiology 35, 247-260