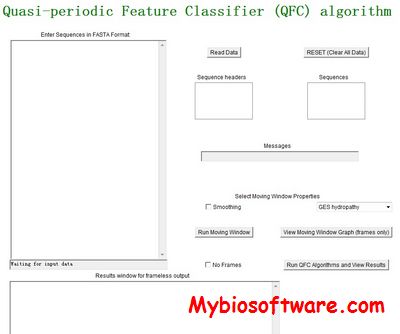
INTREPED 2.0
:: DESCRIPTION
The INTREPED (INTeractive dna REPair prEDiction) web server is a collection of services for predicting properties relating to DNA repair. DNA repair is thought to exist in all organisms that have an active metabolism, and repair systems are crucial for repairing many types of DNA damage. External damage includes (but is not limited to) ionizing (UV) radiation, tobacco smoke, and chemical alteration. Examples of internal damage are DNA copying errors, oxygen byproducts resulting from metabolism, and hydrolysis.
::DEVELOPER
Akutsu Laboratory (Laboratory of Mathematical Bioinformatics)
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Web Browser
:: DOWNLOAD
:: MORE INFORMATION
Citation:
BMC Bioinformatics. 2009 Jan 20;10:25. doi: 10.1186/1471-2105-10-25.
Identification of novel DNA repair proteins via primary sequence, secondary structure, and homology.
Brown JB, Akutsu T.