MAExplorer 0.96.34.01
:: DESCRIPTION
The MAExplorer is a Java-based bioinformatics exploratory data-analysis and data-mining program for analyzing sets of quantitative spotted cDNA or oligonucleotide microarray data . It includes data management, graphics, statistics, clustering, reports, gene data-filtering, user written MAEPlugins, documentation, tutorials, demo data. The exploratory data analysis environment provides tools for the data-mining of quantitative expression profiles across multiple microarrays.
With MAExplorer, it is possible to: 1) analyze the expression of individual genes; 2) analyze the expression of gene families and clusters; 3) compare expression patterns and outliers; 4) directly access other genomic databases for genes of interest. Previously quantified array data is copied to your local computer where it is read by MAExplorer and intermediate results as well as the data mining session state may be saved between data mining sessions.
::DEVELOPER
Dr. Peter Lemkin (LECB/NCI-Frederick) with help from Gregory Thornwall (SAIC) and Jai Evans (DECA/CIT, NIH)
:: SCREENSHOTS

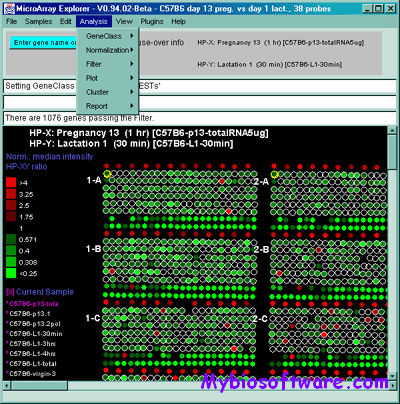
:: REQUIREMENTS
- Windows
- Linux
- MacOsX
- Java
:: DOWNLOAD
 MAExplorer 0.96.34.01 , Manual
MAExplorer 0.96.34.01 , Manual
:: MORE INFORMATION
MAExplorer is available as Open Source at the SourceForge at http://maexplorer.sourceforge.net/ under the Mozilla Public License 1.1 (MPL 1.1) with an acompanying LEGAL document required by the U.S. Government.