CompClust 1.2
:: DESCRIPTION
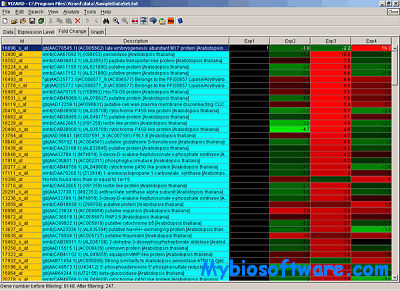
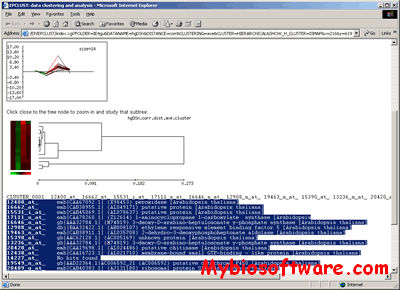
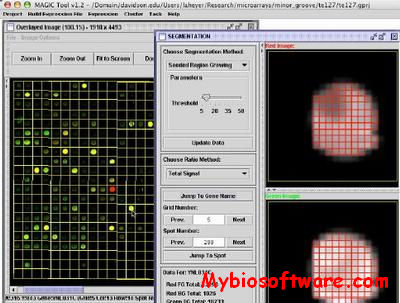
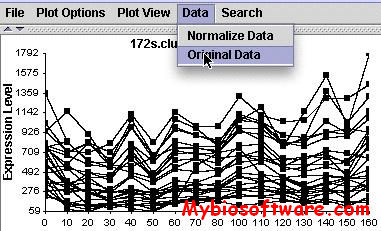
CompClust is a python package written using the pyMLX and IPlot APIs. It provides software tools to explore and quantify relationships between clustering results. Its development has been largely built around needs of microarray data analysis but could be easily used in other domains.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
:: DOWNLOAD
:: MORE INFORMATION
Citation:
Nucleic Acids Res. 2005 May 10;33(8):2580-94. Print 2005.
A mathematical and computational framework for quantitative comparison and integration of large-scale gene expression data.
Hart CE, Sharenbroich L, Bornstein BJ, Trout D, King B, Mjolsness E, Wold BJ.