SBMLsimulator 1.2.1
:: DESCRIPTION
SBMLsimulator is a fast, accurate, and easily usable program for dynamic model simulation and heuristic parameter optimization of models encoded in the Systems Biology Markup Language (SBML). In order to ensure a high reliability of this software, it has been benchmarked against the entire SBML Test Suite and all models from the Biomodels.net database. It includes a large collection of nature-inspired heuristic optimization procedures for efficient model calibration.
::DEVELOPER
the Center for Bioinformatics Tübingen (Zentrum für Bioinformatik Tübingen, ZBIT).
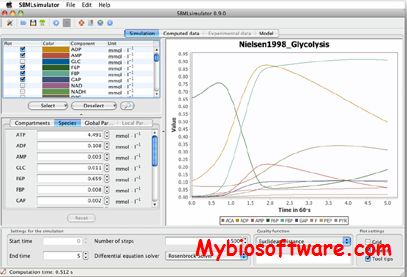
:: SCREENSHOTS
:: REQUIREMENTS
- Linux/ WIndows/MacOsX
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Roland Keller, Alexander Dörr, Akito Tabira, Akira Funahashi, Michael J. Ziller, Richard Adams, Nicolas Rodriguez, Nicolas Le Novère, Noriko Hiroi, Hannes Planatscher, Andreas Zell, and Andreas Dräger.
The systems biology simulation core algorithm.
BMC Systems Biology, 7:55, July 2013